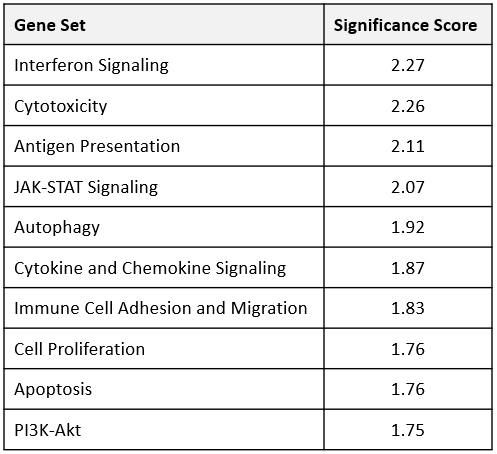
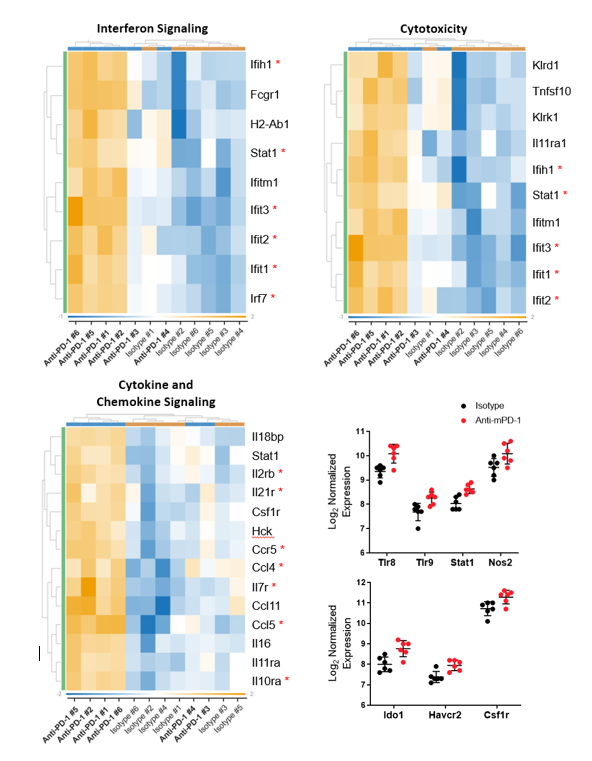
Transcriptomic profiling can be used to examine the interaction between the tumor microenvironment (TME) and the immune system and is a powerful tool to investigate the mechanistic action of immunotherapies. In this Tech Spotlight, we demonstrate how gene expression measured in solid tumors using the nCounter® PanCancer IO 360™ panel (NanoString Technologies) can be combined with flow cytometry to provide a comprehensive evaluation of the effects that immunotherapy has on the anti-tumor immune response.
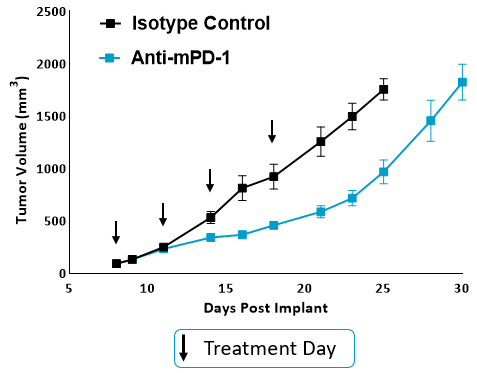
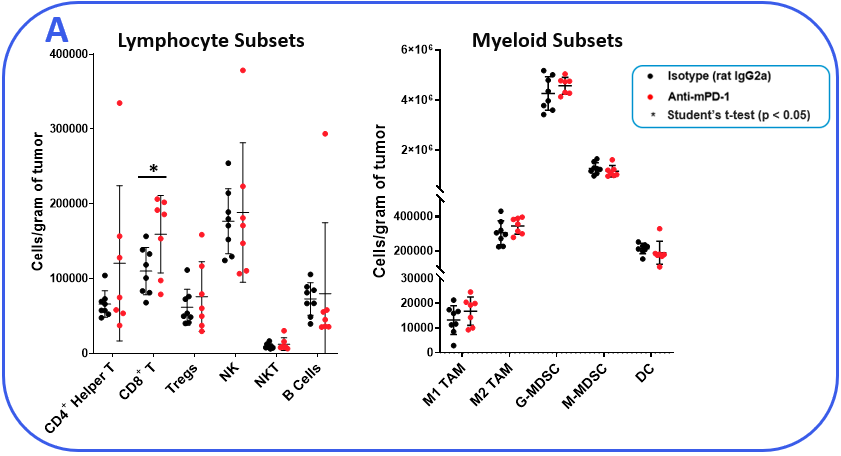
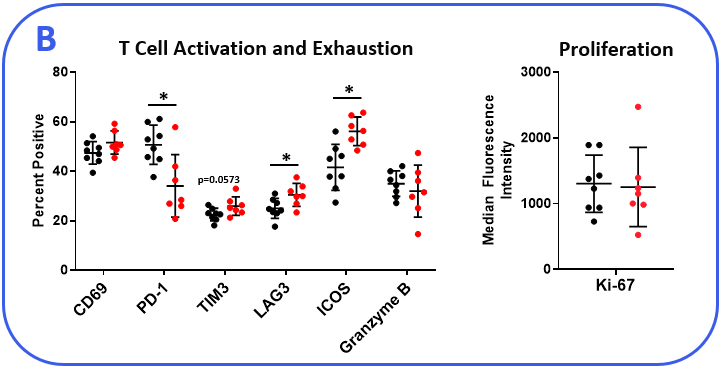
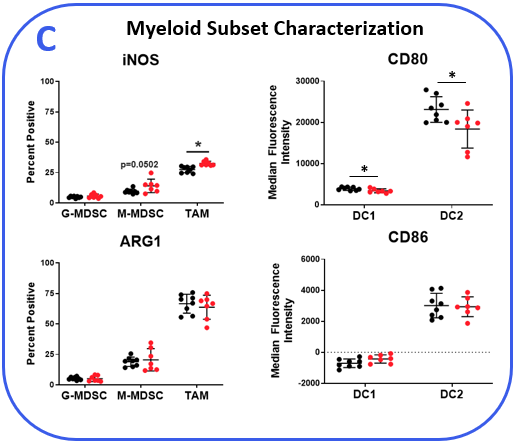
The PanCancer IO 360™ panel is a 770-plex gene expression panel that includes 48 distinct gene signatures to examine cancer-related immune responses. Analysis with the IO 360™ panel provides an unbiased and holistic approach to the examination of the tumor-infiltrating immune cells, tumor cells, and the associated microenvironment. To demonstrate the utility of NanoString analysis when applied to preclinical research, anti-mPD-1 (programmed cell death 1) therapeutic antibody was administered into mice bearing MB49 tumors, which is a murine model for bladder cancer. Flow cytometry was performed to target specific biomarkers for various pro- and anti-tumor activities in different immune subsets. We combined the cytometry and IO 360™ panel results to add an unbiased component to the analysis and provide a global immuno-oncology gene expression profile of the TME in treated versus untreated animals.